ClusterIndex
Quantification of the degree of cell clustering by computing 'Cell Cluster Index'
Description
ClusterIndex is a Jython script for the quantiative evaluation of cell spatial clustering. To measure the degree of cell clustering, we used “nucleus-nucleus distance index (NND)”.
Algorithm
Images with Hoechst labeled nuclei are first binarized using intensity-based automatic image threshold using Otsu algorithm followed by binary fill-hole and open operations. Number of nucleus (\(N_0\)) is then counted using the binary image. Nucleus areas were then dilated by 5 pixels (approximate average diameter of cells for the samples used for the development of this script). If cells were in close proximity to their neighbors and clustered, their nuclei became single object by this dilation, while nuclei of cells remote from others remained as a single object. We counted the number of objects after dilation operation as the cluster number (\(N_5\)) and computed nucleus-nucleus distance index (NND).
NND is computed by dividing the number of cell clusters (\(N_5\)) by the total number of nuclei (\(N_0\)). \[ NND = \frac{N_5}{N_0} \]
NND is close to 1.0 when cells are sparsely distributed while it is close to 0 as cells become clustered.
Dependency
Fiji.
Note that it does not work with pure ImageJ as the script uses auto local threshold function.
Installation
Download the package from the URL linked below, unzip the file and open the script “ClusterIndex.py” in Fiji script editor. See “Workflow” section below for a detailed workflow.
http://dx.doi.org/10.5281/zenodo.18246
Usage
Following is an example workflow. Download the follwoing nucleus image files and open them in Fiji.
Click the image window “cluster.jpg” and bring the image to the front. Then run the script “ClusterIndex.py”.
Click the image window “random.jpg” and bring the image to the front. Then run the script “ClusterIndex.py”
After these two actions, Log window should be showing follwoing measurement results.
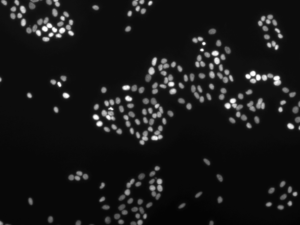
==== cluster.jpg =====
Number of Nucleus : 237
Clusters at dilation 5: 86
Clusters/Nucleus 0.362869198312
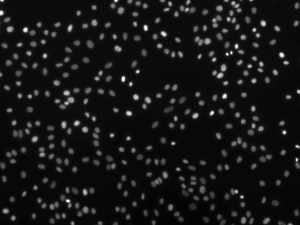
==== random.jpg =====
Number of Nucleus : 346
Clusters at dilation 5: 280
Clusters/Nucleus 0.809248554913
The NND index (“Cluser/Nucleus”) of “cluster.jpg” is 0.36 and that of “random.jpg” is 0.81. This suggest that nucleus in “cluster.jpg” is more clusterd.
Adjusting parameters
To increase / decrease the width of dilation, change the number in line 22 (“SAMPLEITER = 5”). If the radius of cell is 10 pixels, then use that value and change it to “SAMPLEITER = 10”.
To view intermediate images during processing, comment-out line 23 and uncommnet line 24 (“VERBOSE = True”)
Repository
Reference
This script was developed for the analysis reported in the paper below.
Fukuda, S., Nishida-Fukuda, H., Nanba, D., Nakashiro, K., Nakayama, H., Kubota, H., Higashiyama, S., 2016. Reversible interconversion and maintenance of mammary epithelial cell characteristics by the ligand-regulated EGFR system. Sci. Rep. 6, 20209.